Group
Epigenetic Regulation of Cancer Growth and Chemoresistance Group
Unit(s) of assessment: Allied Health Professions, Dentistry, Nursing and Pharmacy
Research theme(s): Health Innovation
School: School of Science and Technology
Overview
Dr Hatziapostolou is an Independent Cancer Researcher who focuses on the molecular interplay between the genome and the epigenome during cancer growth, chemoresistance and metastasis. Cancers of interest are those of the gastrointestinal tract with an emphasis on pancreatic and liver cancer.
Dr Hatziapostolou’s current projects include:
A Systems Biology Approach in Pancreatic Cancer
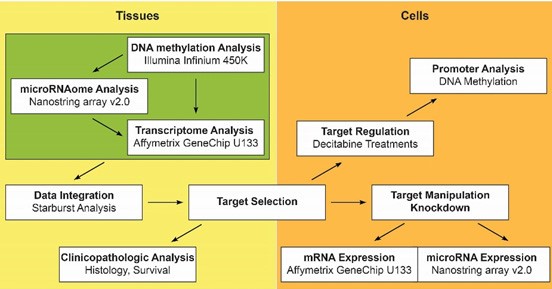
Pancreatic cancer is a complex disease harbouring multiple genetic mutations and widespread epigenetic alterations. The lack of any major improvements in the overall cure rate for pancreatic cancer, the lack of molecular biomarkers for patient selection and the high failure rate of new drugs, in Phase III clinical trials, in pancreatic cancer cells for a more robust pre-clinical and clinical testing of new compounds. The goal of the study is to characterize the molecular interactome of human pancreatic cancer and identify novel druggable pancreatic cancer regulators. By applying genome-wide methylation, transcriptomic and microRNA analyses, this research group has identified an epigenetic circuit that mediates pancreatic epithelial cell transformation, DNA methylation, disease progression and resistance to chemotherapy. This work on pancreatic cancer was recently funded by Pancreatic Cancer Research UK (Research Innovation Award) and attracted the attention of the media. Both Pancreatic Cancer UK and the local press characterised Dr Hatziapostolou’s work as a ‘world-leading research at the cutting edge of science that has the potential to significantly contribute towards the treatment of this deadly disease’.
The Epigenetic Nature of Liver cancer
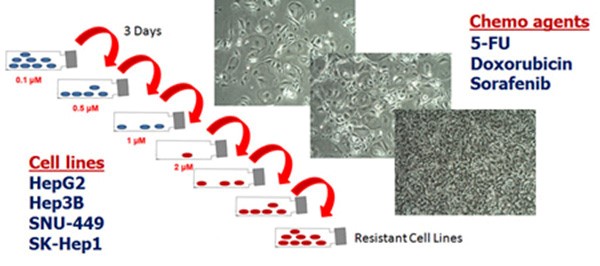
Hepatocellular carcinoma (HCC) represents one of the most frequent visceral and extremely poor prognostic cancers worldwide. Regardless of aetiology, the progression of HCC is highly variable and is influenced by numerous non-genetic factors (age, diet, inflammation), pointing to an epigenetic phenotype. Systemic chemotherapy is being used for advanced disease, with moderate improvement in overall survival duration. These dismal figures point to the development of chemoresistance as an inherent trait of HCC. The research group have established liver cancer cell lines resistant to common chemotherapeutic drugs and performed transcriptomic profiling (for protein-coding genes and non-coding RNAs) and an epigenetic compound screening on the chemoresistant and the parental liver cancer cells. They identified a significant regulatory role of histone demethylases and aim to identify epi-modulator compounds with therapeutic potential, either as a stand-alone regimen or in combination with conventional chemotherapy. The investigation extends to the implication of long non-coding RNAs (LncRNAs) in the regulation of HCC growth and chemoresistance. The group have characterised the role of a specific lncRNA in liver cancer chemoresistance and aim to address its clinical relevance through gene expression profiling in uninvolved and diseased tissues, investigate the functional importance in vivo and explore it they can be therapeutically targeted in HCC.
Transcription Factors in Liver Cancer
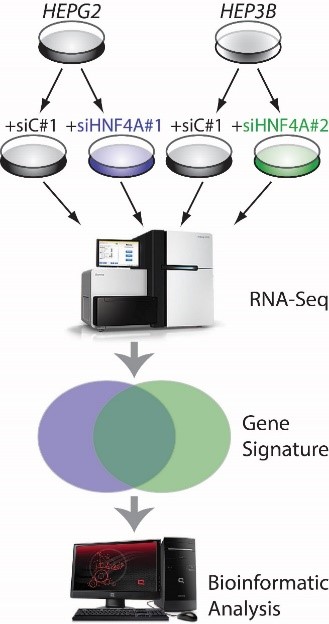
Hepatocyte nuclear factor 4A (HNF4A), is a liver-enriched transcription factor that controls hepatic epithelium formation, hepatocyte differentiation and liver functional activity. The research group have shown (Hatziapostolou et al., Cell, 2011) that HNF4A is a key regulator of hepatocellular carcinoma (HCC) development in murine liver cancer models and human tissues. The goal of this project is to elucidate the role of HNF4A-regulated gene networks in liver cancer chemoresistance and metastasis and identify new therapeutic schemes that could potentially target both the dissemination of cancer cells and drug resistance.
Collaboration
Dr Hatziapostolou collaborates with top researchers in the field of cancer from a number of universities across the UK, Europe and USA.
Membership
Publications
H3K4me3 regulates glucose metabolism and lipid production through KMT2D histone methyltransferase in pancreatic cancer. Koutsioumpa M, Hatziapostolou M, Polytarchou C, Mahurkar-Joshi S, Williams J, Tirado-Rodriguez B, Huerta-Yepez S, Karavias D, Kourea H, Poultsides GA, Dawson DW, Donahue TR, Iliopoulos D. (2019). Gut. Jul;68(7):1271-1286.
Transcriptomic and CRISPR/Cas9 technologies reveal FOXA2 as a tumour suppressor gene in pancreatic cancer. Vorvis C, Hatziapostolou M, Mahurkar-Joshi S, Koutsioumpa M, Williams J, Donahue TR, Poultsides GA, Eibl G, Iliopoulos D. (2016). Am. J. Physiol. Gastrointest. Liver Physiol. 310(11):G1124-37.
XBP1 promotes triple negative breast cancer by controlling the HIF1A pathway. Chen X, Iliopoulos D#, Zhang Q#, Tang Q#, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, Mai J, Shen H, Hu DZ, Adoro S, Hu B, Song M, Tan C, Landis MD, Ferrari M, Shin SJ, Brown M, Chang JC, Liu XS, Glimcher LH. (2014). Nature. 508(7494):103-7. (#equal contribution).
Identification of liver cancer progenitors whose malignant progression depends on IL-6 signaling. He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, Shalapour S, Seki E, Yost SE, Jepsen K, Frazer KA, Harismendy O, Hatziapostolou M, Iliopoulos D, Suetsugu A, Hoffman RM, Tateishi R, Koike K, Karin M. (2013). Cell. 155(2):384-96.
An HNF4A-microRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, Iliopoulos D. (2011). Cell. 147(6):1233-47.
Tumour progression locus 2 regulates signal-induced calcium release from the endoplasmic reticulum and cell migration. Hatziapostolou M, Koukos G, Polytarchou C, Serebrennikova O, Kuliopulos A, Tsichlis PN. Science Signaling. (2011). 4(187):ra55.
Facilities
The laboratories of Dr Hatziapostolou house the following equipment:
1. S220 Focused-Ultrasonicator from Covaris: delivers controlled energy precisely and accurately to sample volumes ranging from 15 μl to 18 ml.
2. CFX384 Real-Time PCR detection system from Bio-Rad: rapid data acquisition, ideal for performing high-throughput real-time PCR in a 384-well format.
3. Bio-Plex Multiplex Immunoassay System from Bio-Rad: Quantification of over 500 different protein and peptide targets simultaneously in a single sample. High-throughput and sensitive assays enabling the acquisition of high-quality data.
We also have access within the John van Geest Cancer Research Centre to instrumentation to facilitate multi-omic analyses including gene expression micro-array, Nanostring platform, mass spectrometers and laser capture microdissection.